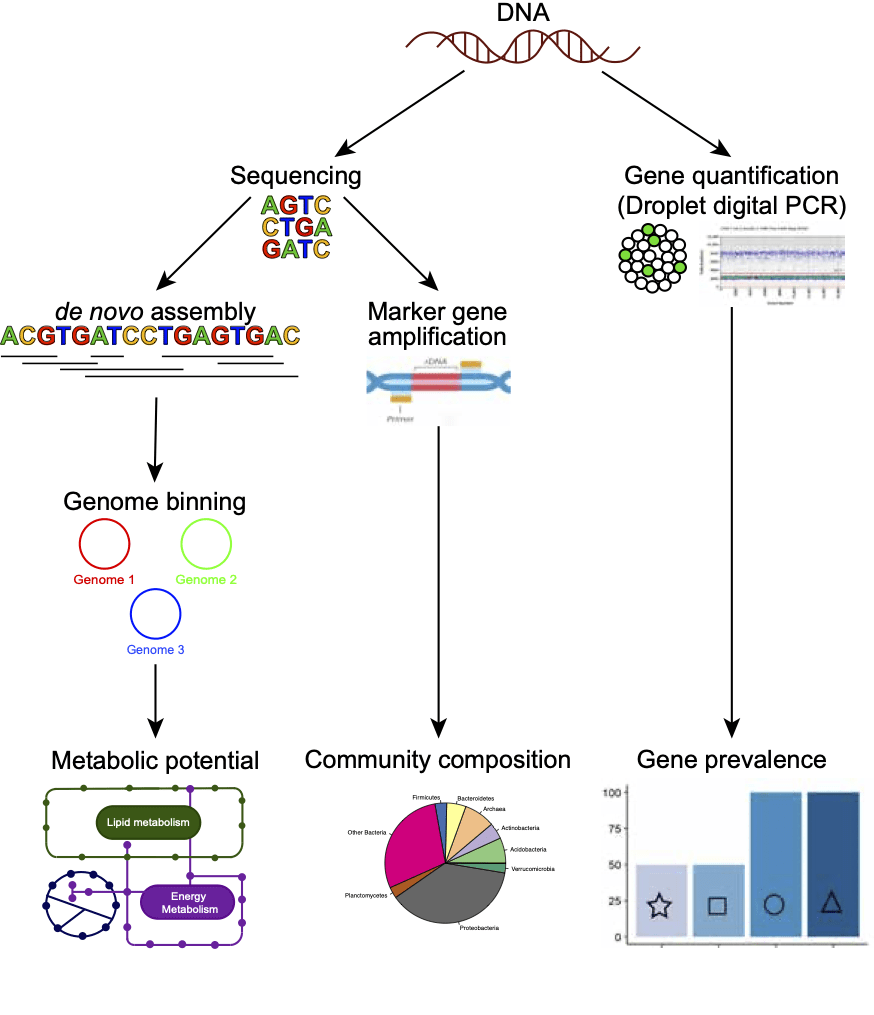
Genes & genomes (DNA-based approaches)
Genome assembly: We sequenced the whole DNA present in groundwater samples, and then used this to assemble hundreds of microbial genomes. By analysing the genes present we can determine what these organisms are capable of metabolically, including determining their capacity (or metabolic potential) for nitrogen removal.
Taxonomic marker genes: We also targeted and amplified taxonomic marker genes (via polymerase chain reaction, PCR) in prokaryotes (bacteria and archaea) and eukaryotes (microeukaryotes and stygofauna or subterranean animals) before sequencing these. Sequencing these marker genes enabled us to determine the identity of organisms present in the groundwater, including rarer organisms not recovered by genomics. Taken together, this approach provides information on the composition of groundwater communities.
Gene quantification:
Fluorescence was used to measure the concentration of metabolic marker genes (amplified during droplet digital PCR) in groundwater. This approach was used to quantify genes available for critical steps in the microbial nitrogen cycle, and determine their prevalence across New Zealand aquifers.
These approaches enable us to:
Determine the metabolic potential present in groundwater microorganisms
Determine the composition of organisms present in groundwater (microbial and eukaryotic)
Quantify key genes in the microbial nitrogen cycle