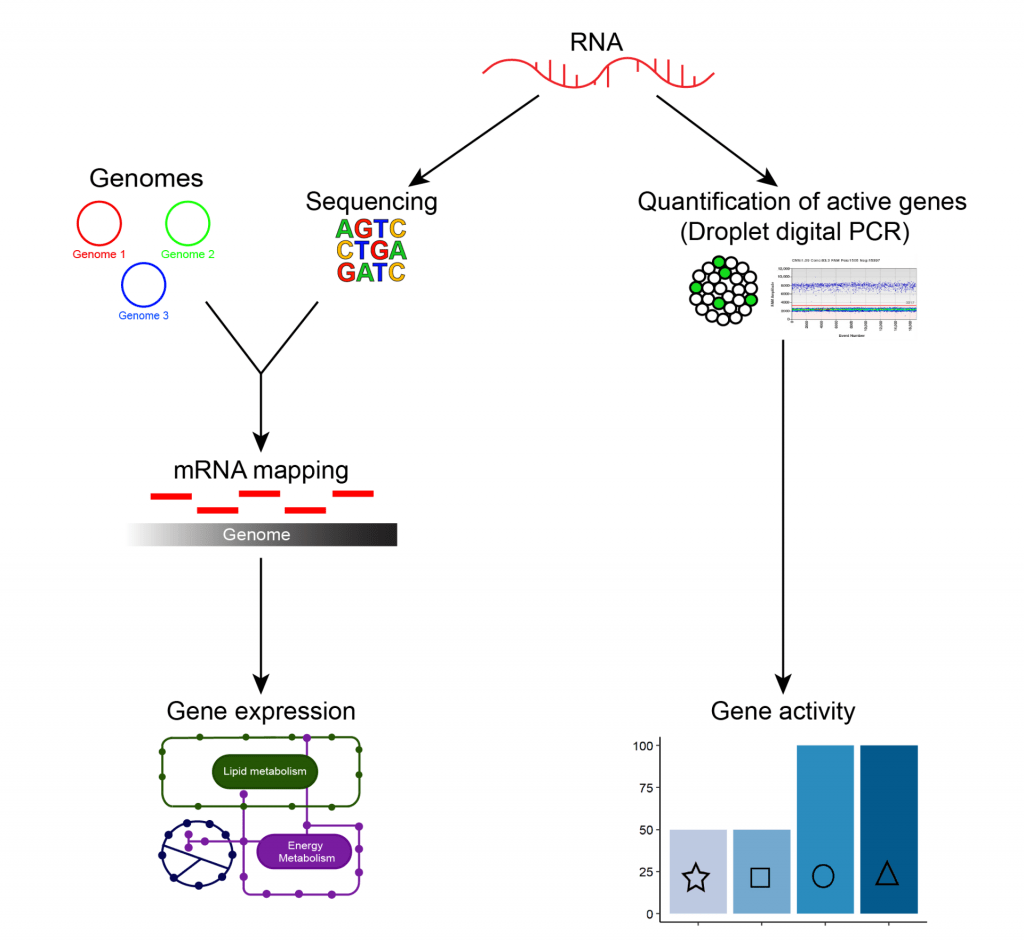
Gene expression (RNA-based approaches)
RNA Sequencing:
We sequenced the RNA present in groundwater samples, and then mapped this back to our database of microbial genomes from the same groundwater. This enables us to determine which genes are being actively used (transcribed into messenger RNA, or mRNA, which is in turn used in the synthesis of proteins), and by which organisms. Ultimately we can use this information to determine what groundwater microorganisms are doing, individually and collectively, including how they are transforming nutrients (and potential contaminants), such as nitrogen.
Gene transcript quantification:
As for gene quantification, fluorescence was used to measure the concentration of metabolic marker gene transcripts (amplified during droplet digital PCR) in groundwater. This approach was used to quantify genes being actively transcribed to undertake critical steps in the microbial nitrogen cycle.
These approaches enable us to:
Determine the metabolic activity of organisms present in groundwater
Quantify the activity of key genes in the microbial nitrogen cycle